[link]
Summary by Tiago Vinhoza 7 years ago
#### Goal
+ Use unsupervised deep learning to obtain a low-dimensional representation of a patient from EHR data.
+ A better representation will facilitate clinical prediction tasks.
#### Architecture:
+ Patient EHR is obtained from the Hospital Data Warehouse:
+ demographic info
+ ICD-9 codes
+ medication, labs
+ clinical notes: free text
+ Use stacked denoising autoencoders (SDA) to obtain an abstract representation of the patient with lower dimensionality.
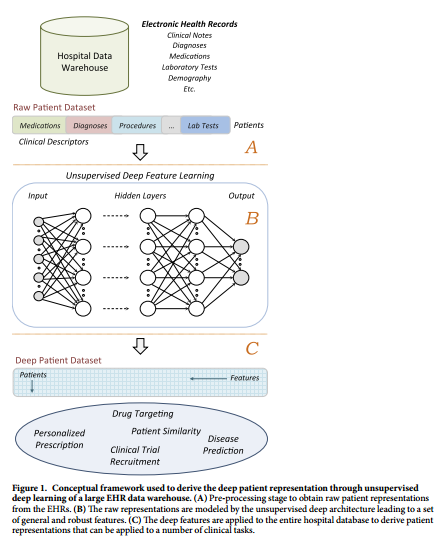
#### Dataset:
+ Data Warehouse from Mount Sinai Hospital in NY.
+ All patient records that had a diagnosed disease (ICD-9 code) between 1980 and 2014 - approximately 1.2 million patients with 88.9 records/patient - were initially selected.
+ 1980-2013: training, 2014: test.
*Data Cleaning*:
+ Diseases diagnosed in fewer than 10 patients in the training dataset were eliminated.
+ Diseases that could not be diagnosed through EHR labels were eliminated. Related to social behavior (HIV), fortuitous events (injuries, poisoning) or unspecific ('other cancers'). The final list contains 78 diseases.
*Final version of the dataset (raw patient representation)*:
+ Training: 704,587 patients (to obtain deep features post SDA).
+ Validation: 5,000 patients (for the evaluation of the predictive model for diseases).
+ Test: 76,214 patients (for the evaluation of the predictive model for diseases).
+ 41072 columns - demographic info, ICD-9, medication, lab test, free text (LDA topic modeling dimension 300)
+ Very high dimensional but very sparse representation
#### Results:
*Stacked Denoisinig Autoencoders for low-dimensional patient representation*:
+ 3 layers of denoising autoencoders.
+ Each layer has 500 neurons. Patient is now represented by a dense vector of 500 features.
+ Inputs are normalized to lie in the [0, 1] interval.
+ Inputs in each of the layers have added noise at a ratio of 5% noise (masking noise corruption - value of these features is set to '0').
+ Sigmoid activation function.
*Classifiers for disease prediction*:
+ Random forest classifiers with 100 trees trained for each of the 78 diseases.
*Baseline for comparison*:
+ PCA with 100 components, k-means with 500 clusters, GMM with 200 mixes and ICA with 100 components. (see Discussion)
+ RawFeat: original patient EHR features: sparse vector with 41072 features (~ 1% of non-zero entries).
+ Threshold to rank as "positive": 0.6
*Aggregate performance in predicting diseases*:
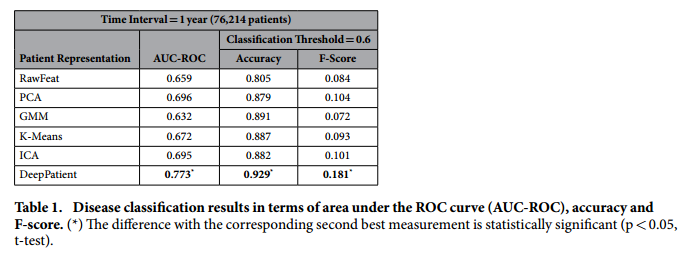
+ Comment: This result of F-Score = 0.181 implies a precision of 0.102 (let us assume a recall in the order of 80%), which means that with each correct diagnosis, the Deep Patient generates approximately 9 false alarms.
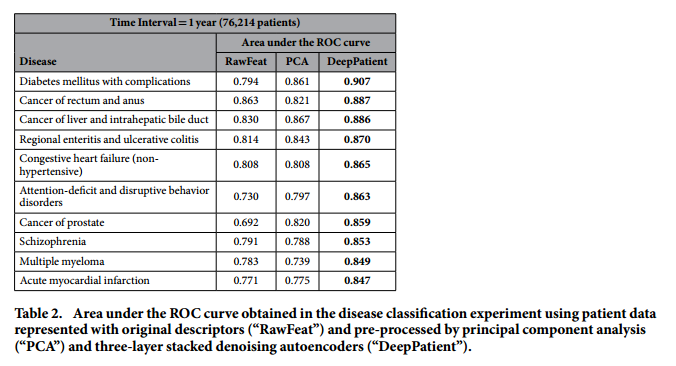
*Performance for some particular diseases*:
#### Discussion:
+ DeepPatient *does not* use lab results in model building. Only the *frequency* at which the analysis is performed is taken into account.
+ Future enhancements:
+ Describe a patient with a temporal sequence of vectors s instead of summarizing all data in one vector.
+ Add other categories of EHR data, such as insurance details, family history and social behaviors.
+ Use PCA as a pre-processing step before SDA?
+ Caveat: the comparisons does not seem to be fair. If the autoencoder has dimension 500, the other baselines should also have dimension 500.














