[link]
Summary by Tiago Vinhoza 8 years ago
#### Motivation:
+ Take advantage of the fact that missing values can be very informative about the label.
+ Sampling a time series generates many missing values.

#### Model (indicator flag):
+ Indicator of occurrence of missing value.
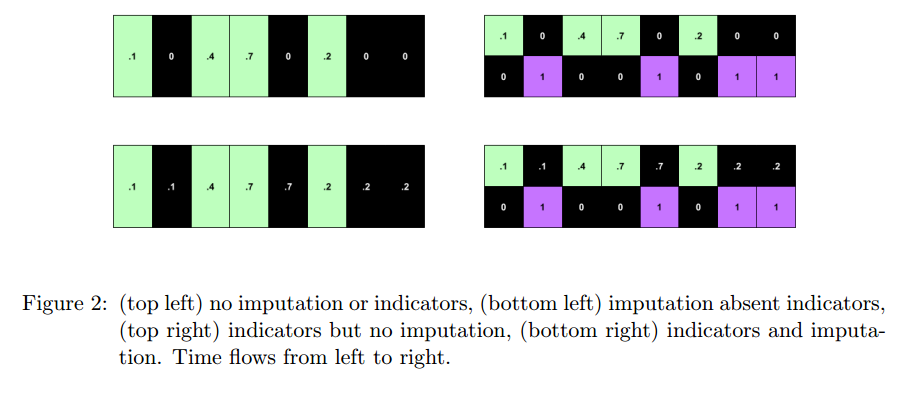
+ An RNN can learn about missing values and their importance only by using the indicator function. The nonlinearity from this type of model helps capturing these dependencies.
+ If one wants to use a linear model, feature engineering is needed to overcome its limitations.
+ indicator for whether a variable was measured at all
+ mean and standard deviation of the indicator
+ frequency with which a variable switches from measured to missing and vice-versa.
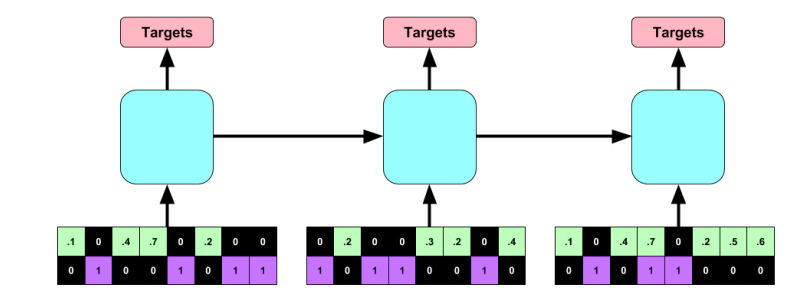
#### Architecture:
+ RNN with target replication following the work "Learning to Diagnose with LSTM Recurrent Neural Networks" by the same authors.
#### Dataset:
+ Children's Hospital LA
+ Episode is a multivariate time series that describes the stay of one patient in the intensive care unit
Dataset properties | Value
---------|----------
Number of episodes | 10,401
Duration of episodes | From 12h to several months
Time series variables | Systolic blood pressure, Diastolic blood pressure, Peripheral capillary refill rate, End tidal CO2, Fraction of inspired O2, Glasgow coma scale, Blood glucose, Heart rate, pH, Respiratory rate, Blood O2 Saturation, Body temperature, Urine output.
#### Experiments and Results:
**Goal**
+ Predict 128 diagnoses.
+ Multilabel: patients can have more than one diagnose.
**Methodology**
+ Split: 80% training, 10% validation, 10% test.
+ Normalized data to be in the range [0,1].
+ LSTM RNN:
+ 2 hidden layers with 128 cells. Dropout = 0.5, L2-regularization: 1e-6
+ Training for 100 epochs. Parameters chosen correspond to the time that generated the smallest error in the validation dataset.
+ Baselines:
+ Logistic Regression (L2 regularization)
+ MLP with 3 hidden layers and 500 hidden neurons / layer (parameters chosen via validation set)
+ Tested with raw-features and hand-engineered features.
+ Strategies for missing values:
+ Zeroing
+ Impute via forward / backfilling
+ Impute with zeros and use indicator function
+ Impute via forward / backfilling and use indicator function
+ Use indicator function only
#### Results
+ Metrics:
+ Micro AUC, Micro F1: calculated by adding the TPs, FPs, TNs and FNs for the entire dataset and for all classes.
+ Macro AUC, Macro F1: Arithmetic mean of AUCs and F1 scores for each of the classes.
+ Precision at 10: Fraction of correct diagnostics among the top 10 predictions of the model.
+ The upper bound for precision at 10 is 0.2281 since in the test set there are on average 2.281 diagnoses per patient.

#### Discussion:
+ Predictive model based on data collected following a given routine. This routine can change if the model is put into practice. Will the model predictions in this new routine remain valid?
+ Missing values in a way give an indication of the type of treatment being followed.
+ Trade-off between complex models operating on raw features and very complex features operating on more interpretable models.














